Lipid Rafts and a Possible Cure for Cancer That Could Go Viral
Every discipline, every area of expertise, has its controversial topics. The lipid specialists are no exception. The topic that divides them is the theory of lipid rafts.
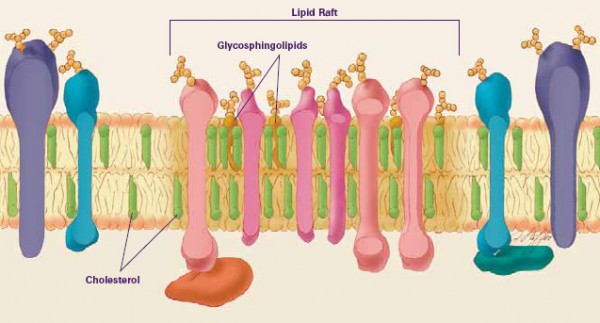
Schematic of a lipid raft, from the perspective of a cross-section of the cellular membrane. Diagram from the (American) National Institute of General Medical Sciences.
Since it became clear that a variety of lipids are present in cellular membranes, investigations about how these are arranged have been undertaken. One face of this approach is to understand the full gamut of lipids in the system (lipidomics), and the other is a physical analysis of the behaviour of the system. These two angles of understanding have increased in depth, with physical studies looking to solve the problem using a variety of experiments, many of which are based on X-ray scattering [1-3] or Nuclear Magnetic Resonance (NMR) [2,4]. These sorts of studies have found a variety of complexities in biological systems comprising a large number of lipid components that are not observed in model systems, that typically comprise many fewer). Some of these variations have been identified as inhomogeneities in the membranes under study. Such inhomogeneities are often observed by variations in the phase behaviour of smaller parts of the system. This concept is taken a stage further with the theory of lipid rafts in that there are small, local areas that have a dissimilar lipid composition to the membrane that surrounds it, leading to the different phase behaviour. There is also evidence that cholesterol in the membrane can influence this phase behaviour [2].
The idea of small areas of different lipid composition to the cellular average may sound unremarkable, but it can also be used to explain differences in protein activity with respect to lipid composition [5]. Certainly, there are examples of changes to protein activity as a result of changes in lipid composition that cannot be explained on purely biological grounds [6]. Despite that, data from a recent biological study in cancerous cells indicates that certain target proteins are located in lipid rafts [5]. One such protein is called prohibitin (PHB). This is activated by another protein, a kinase called PI3K. This is a protein responsible for installing a phosphate group – a process called phosphorylation. However, the effect of this PI3K can be suppressed by the presence of a protein called rVP1. This is especially useful, because it has been shown that application of this protein induces cell death (apoptosis) in cervical cancer cells.
rVP1 was first discovered from a seemingly unlikely source: it is one of the four types of protein found in the Food and Mouth Disease virus, Aphthovirus. Its precise action in suppressing phosphorylation of PHB is unclear, however there is evidence that it reduces the amount of phosphatidylinositol-3,4,5-tris-phosphate (PIP3, another lipid), an important signalling lipid that requires three phosphorylations to be produced from phosphatidylinositol. Evidence also suggests that rVPI can reduce the phosphorylation-mediated activity of other proteins in vivo [5]. Additionally, changes to the inositide profile of the membrane’s lipid fraction may have a purely physical influence on the behaviour of that membrane [3, 7].
The study by Chiu et al. [5] suggests there is a complicated set of interactions between lipids and proteins, both in biological terms, though signalling mechanisms, and by the physical behaviour of the systems. rVP1’s action can easily be seen as an exciting possibility for cancer therapy, but also as a note of caution when we interpret the data as showing that a single molecule (rVP1) can have serious effects, both biologically and physically, and that occur simultaneously.
References
[1] F. Evers, C. Jeworrek, K. Weise , M. Tolan, R. Winter, Soft Matter, 2012, 8, 2170-2175.
[2] D. L. Gater, J. M. Seddon, R. V. Law, Soft Matter, 2008, 4, 263-267.
[3] S. Furse, N. J. Brooks, A. M. Seddon, R. Woscholski, R. H. Templer, E. W. Tate, P. R. J. Gaffney, O. Ces, Soft Matter, 2012, 8, 3090-3093.
[4] J. A. Clarke, J. M. Seddon, R. V. Law, Soft Matter, 2009, 5, 369-378.
[5] C. F. Chiu, J. M. Peng, S. W. Hung, C. M. Liang, S. M. Liang, Cancer Letters, 2012, 320, 205-214.
[6] M. Younsi, D. Quilliot, N. Al-Makdissy, I. Delbachian, P. Drouin, M. Donner, O. Ziegler; Metabolism, 2002, 51, 1261-1268.
[7] X. Mulet, R. H. Templer, R. Woscholski, O. Ces, Langmuir, 2008, 24 , 8443–8447. http://pubs.acs.org/doi/abs/10.1021/la801114n.